Image-based mushroom identification
Local Processing:
Remote & Notebook Processing:
Artifacts:
| Dataset: | images.tgz | train.tgz | test.tgz |
|---|---|---|---|
| Annotator json: | images.json | categories.json | config.json |
| @ai.columbari.us: | Web Annotator! | mo_example_task.tar.gz | mo_example_task.zip |
Setup (Locally)
# venv:
python3 -m venv mushroomobserver_venv
source mushroomobserver_venv/bin/activate
pip3 install -r requirements.txt
preprocess: (Locally)
python3 preprocess
- Fetches & saves off gbif archive to
./static/- Checks the archive, tries loading it into memory etc
- Fetches Leaflet Annotator binary & licenses from JessSullivan/MerlinAI-Interpreters
- Generates an
images.jsonannotation file from the assets selected by Joe & Nathan - Generates an
categories.jsonfile from the annotatable classes selected by Joe & Nathan - Downloads, organizes the selected assets from images.mushroomoberver.org at
./static/images/<category>/<id>.jpg- writes out images archive
- More or less randomly divvies up testing & training image sets
- writes out example testing/training archives; (while training it’ll probably be easier to resample directly from images.tgz from keras)
Structures:
- Leaflet Annotator
images.jsonStructure:- id: filename The MO image filename
- category_id: The binomen defined in the
./static/sample_select_assets.csv; for directories and URIs this is converted to snake case. - taxon_id: the MO taxon id integer in the case of
*_v1.csv; otherwise duplicate ofcategory_idfor now (eg*_v2.csv) - url: Temporary elastic ip address this asset will be available from, just to reduce any excessive / redundant traffic to images.mushroomobserver.org
- src: imageURL The asset’s source URL form Mushroom Observer
[{ "id": "290214.jpg", "taxon_id": "12326", "category_id": "Peltula euploca", "url": "https://mo.columbari.us/static/images/peltula_euploca/290214.jpg", "src": "https://mushroomobserver.org/images/640/290214.jpg" }]
./static/directory structure followingpython3 preprocess:├── static ├── categories.json ├── gbif.zip ├── images | ... │ └── peltula_euploca │ ├── <123456>.jpg │ ... │ └── <654321>.jpg │ ... ├── images.tgz ├── testing | ... │ └── peltula_euploca │ ├── <234567>.jpg │ ... │ └── <765432>.jpg │ ... ├── testing.tgz ├── training | ... │ └── peltula_euploca │ ├── <345678>.jpg │ ... │ └── <876543>.jpg │ ... ├── images.json ├── training.tgz ├── js │ ├── leaflet.annotation.js │ └── leaflet.annotation.js.LICENSE.txt ├── md │ ├── output_10_0.png │ ├── output_11_0.png │ ├── output_17_0.png │ └── output_27_0.png ├── mo_example_task │ ├── categories.json │ ├── config.json │ └── images.json ├── sample_select_assets_v1.csv ├── sample_select_assets_v2.csv ├── test.tgz └── train.tgz ...
Train (Locally)
python3 train
- Fetches, divvies & shuffles train / validation sets from within Keras using archive available at mo.columbari.us/static/images.tgz
- More or less running Google’s demo transfer learning training script in
train/training_v1.pyas of 03/17/21, still need to bring in training operations and whatnot from merlin_ai/ repo –> experiment with Danish Mycology Society’s ImageNet v4 notes
### Local Jupyter:
- One may also open and run notebooks locally like this:
- rename ipython notebook:
cp train/notebook/training_v1.ipynb.bak train/notebook/training_v1.ipynb - launch jupyter:
jupyter notebook - or without authentication:
jupyter notebook --ip='*' --NotebookApp.token='' --NotebookApp.password=''
- rename ipython notebook:
### Google Colab:
- @gvanhorn38 pointed out Google Colabs’s neat Juptyer notebook service will train models for free if things are small enough- I have no idea what the limits are- fiddle with their intro to image classification on Google Colab here, its super cool!
Notes:
## Preprocess: (Jupyter)
from load_dwca import MODwca
from preprocess import Preprocess
from common import *
"""MODwca():
fetch & save off the gbif export
make sure we can load the dwca archive into memory:
"""
dwca = MODwca()
"""BuildImages():
functions to construct image dataset and annotator artefacts
"""
buildData = Preprocess()
buildData.write_images_json()
buildData.fetch_leaflet_tool()
buildData.fetch_online_images(_json=STATIC_PATH + "images.json")
buildData.export_tgz()
buildData.split_training_testing()
buildData.write_categories_json()
## Train: (Jupyter)
import matplotlib.pyplot as plt
import numpy as np
import os
import PIL
import tensorflow as tf
import pathlib
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
dataset_url = "https://mo.columbari.us/static/images.tgz"
data_dir = tf.keras.utils.get_file('', origin=dataset_url, untar=True)
data_dir = pathlib.Path(data_dir)
image_count = len(list(data_dir.glob('*/*.jpg')))
print("Read " + str(image_count) + " images into Keras!")
armillaria_tabescens = list(data_dir.glob('armillaria_tabescens/*'))
PIL.Image.open(str(armillaria_tabescens[0]))

PIL.Image.open(str(armillaria_tabescens[1]))

load parameters:
batch_size = 32
img_height = 500
img_width = 375
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.1,
subset="training",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size)
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.1,
subset="validation",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size)
class_names = train_ds.class_names
num_classes = len(class_names)
print("Read " + str(num_classes) + " classification classes:")
x=1
for _name in class_names:
print(str(x) + ": " + _name)
x += 1
## Visualize:
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 10))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(class_names[labels[i]])
plt.axis("off")

for image_batch, labels_batch in train_ds:
print("Processed image batch shape: " + image_batch.shape.__str__())
print("Processed labels batch shape: " + labels_batch.shape.__str__())
break
AUTOTUNE = tf.data.AUTOTUNE
train_ds = train_ds.cache().shuffle(1000).prefetch(buffer_size=AUTOTUNE)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
normalization_layer = layers.experimental.preprocessing.Rescaling(1./255)
normalized_ds = train_ds.map(lambda x, y: (normalization_layer(x), y))
image_batch, labels_batch = next(iter(normalized_ds))
first_image = image_batch[0]
# Notice the pixels values are now in `[0,1]`.
print(np.min(first_image), np.max(first_image))
data_augmentation = keras.Sequential(
[
layers.experimental.preprocessing.RandomFlip("horizontal",
input_shape=(img_height,
img_width,
3)),
layers.experimental.preprocessing.RandomRotation(0.1),
layers.experimental.preprocessing.RandomZoom(0.1),
]
)
model = Sequential([
data_augmentation,
layers.experimental.preprocessing.Rescaling(1./255),
layers.Conv2D(16, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(32, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(64, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Dropout(0.2),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dense(num_classes)
])
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
model.summary()
epochs = 15
history = model.fit(
train_ds,
validation_data=val_ds,
epochs=epochs
)
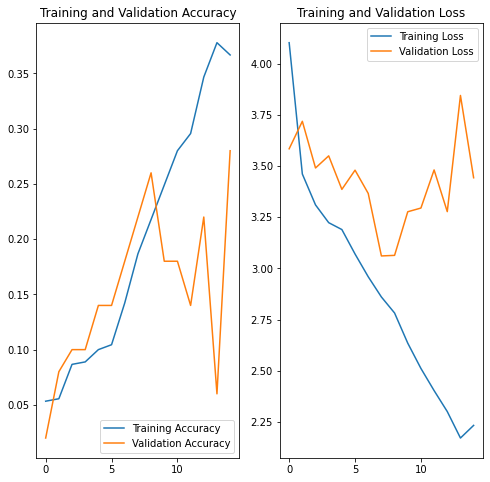
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs_range = range(epochs)
plt.figure(figsize=(8, 8))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label='Training Accuracy')
plt.plot(epochs_range, val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.title('Training and Validation Accuracy')
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.title('Training and Validation Loss')
plt.show()
## Predict:
tabescens_url = "https://www.mushroomexpert.com/images/kuo6/armillaria_tabescens_06.jpg"
tabescens_path = tf.keras.utils.get_file('armillaria_tabescens_06', origin=tabescens_url)
img = keras.preprocessing.image.load_img(
tabescens_path, target_size=(img_height, img_width)
)
img_array = keras.preprocessing.image.img_to_array(img)
img_array = tf.expand_dims(img_array, 0)
predictions = model.predict(img_array)
score = tf.nn.softmax(predictions[0])
print(
"This image most likely belongs to {} with a {:.2f} percent confidence."
.format(class_names[np.argmax(score)], 100 * np.max(score))
)
This image most likely belongs to armillaria_tabescens with a 45.49 percent confidence.